This vignette contains a number of examples on how to use
clugenr in 2D. Examples require the following setup
code:
# Load the clugenr library
library(clugenr)
# Load functions for plotting examples
source("plot_examples_2d.R", local = knitr::knit_global())
# Keep examples reproducible in newer R versions
RNGversion("3.6.0")The 2D examples were plotted with the plot_examples_2d()
function available here.
Manipulating the direction of cluster-supporting lines
Using the direction parameter
seed <- 123
e001 <- clugen(2, 4, 200, c(1, 0), 0, c(10, 10), 10, 1.5, 0.5, seed = seed)
e002 <- clugen(2, 4, 200, c(1, 1), 0, c(10, 10), 10, 1.5, 0.5, seed = seed)
e003 <- clugen(2, 4, 200, c(0, 1), 0, c(10, 10), 10, 1.5, 0.5, seed = seed)
plot_examples_2d(list(e = e001, t = "e001: direction = [1, 0]"),
list(e = e002, t = "e002: direction = [1, 1]"),
list(e = e003, t = "e003: direction = [0, 1]"))
Changing the angle_disp parameter and using a custom
angle_deltas_fn function
seed <- 123
# Custom angle_deltas function: arbitrarily rotate some clusters by 90 degrees
angdel_90_fn <- function(nclu, astd) sample(c(0, pi / 2), nclu, replace = TRUE)
e004 <- clugen(2, 6, 500, c(1, 0), 0, c(10, 10), 10, 1.5, 0.5, seed = seed)
e005 <- clugen(2, 6, 500, c(1, 0), pi / 8, c(10, 10), 10, 1.5, 0.5, seed = seed)
e006 <- clugen(2, 6, 500, c(1, 0), 0, c(10, 10), 10, 1.5, 0.5, seed = seed,
angle_deltas_fn = angdel_90_fn)
plot_examples_2d(list(e = e004, t = "e004: angle_disp = 0"),
list(e = e005, t = "e005: angle_disp = π/8"),
list(e = e006, t = "e006: custom angle_deltas function"))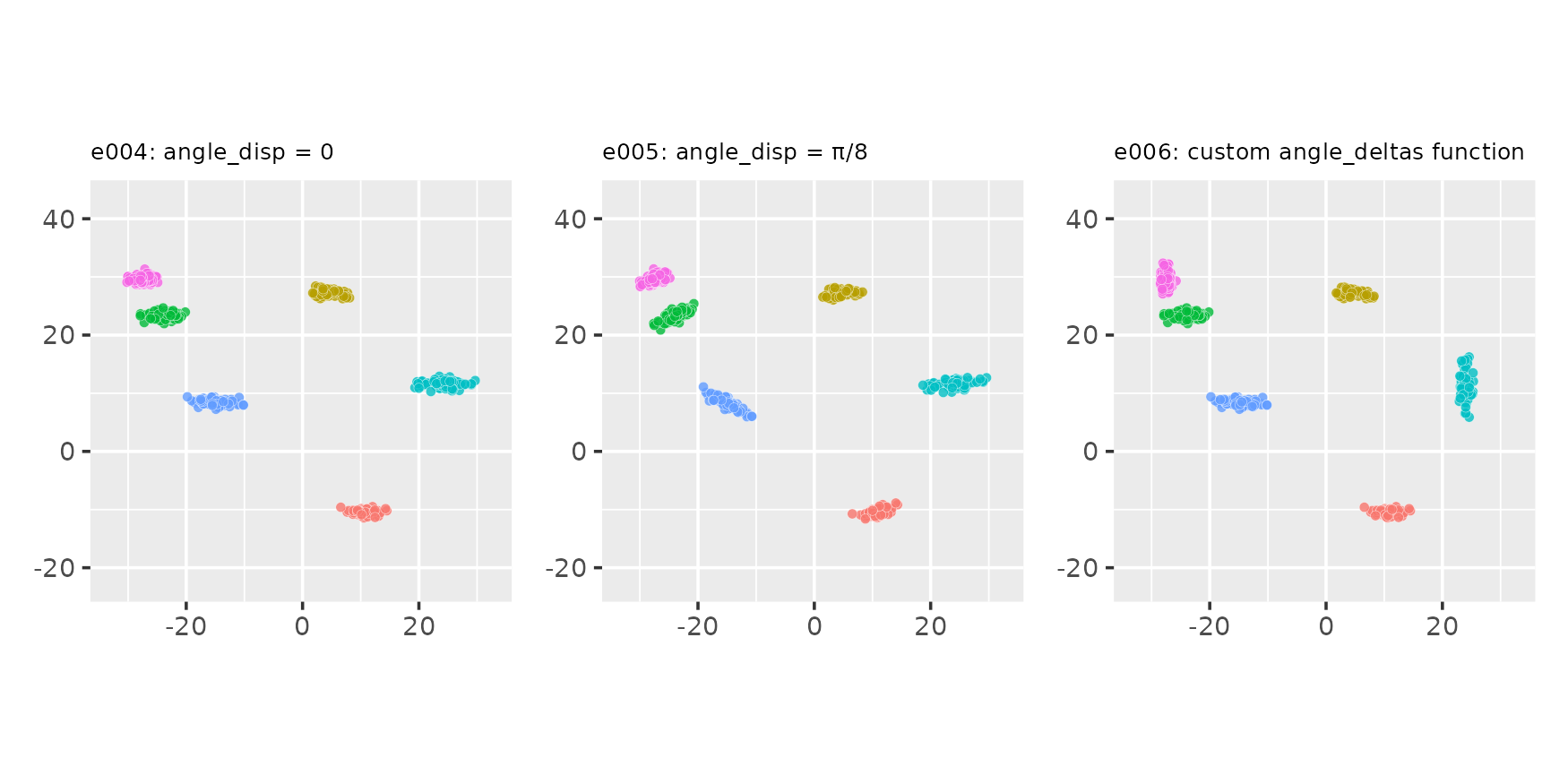
Manipulating the length of cluster-supporting lines
Using the llength parameter
seed <- 321
e007 <- clugen(2, 5, 800, c(1, 0), pi / 10, c(10, 10), 0, 0, 0.5, seed = seed,
point_dist_fn = "n")
e008 <- clugen(2, 5, 800, c(1, 0), pi / 10, c(10, 10), 10, 0, 0.5, seed = seed,
point_dist_fn = "n")
e009 <- clugen(2, 5, 800, c(1, 0), pi / 10, c(10, 10), 30, 0, 0.5, seed = seed,
point_dist_fn = "n")
plot_examples_2d(list(e = e007, t = "e007: llength = 0"),
list(e = e008, t = "e008: llength = 10"),
list(e = e009, t = "e009: llength = 30"))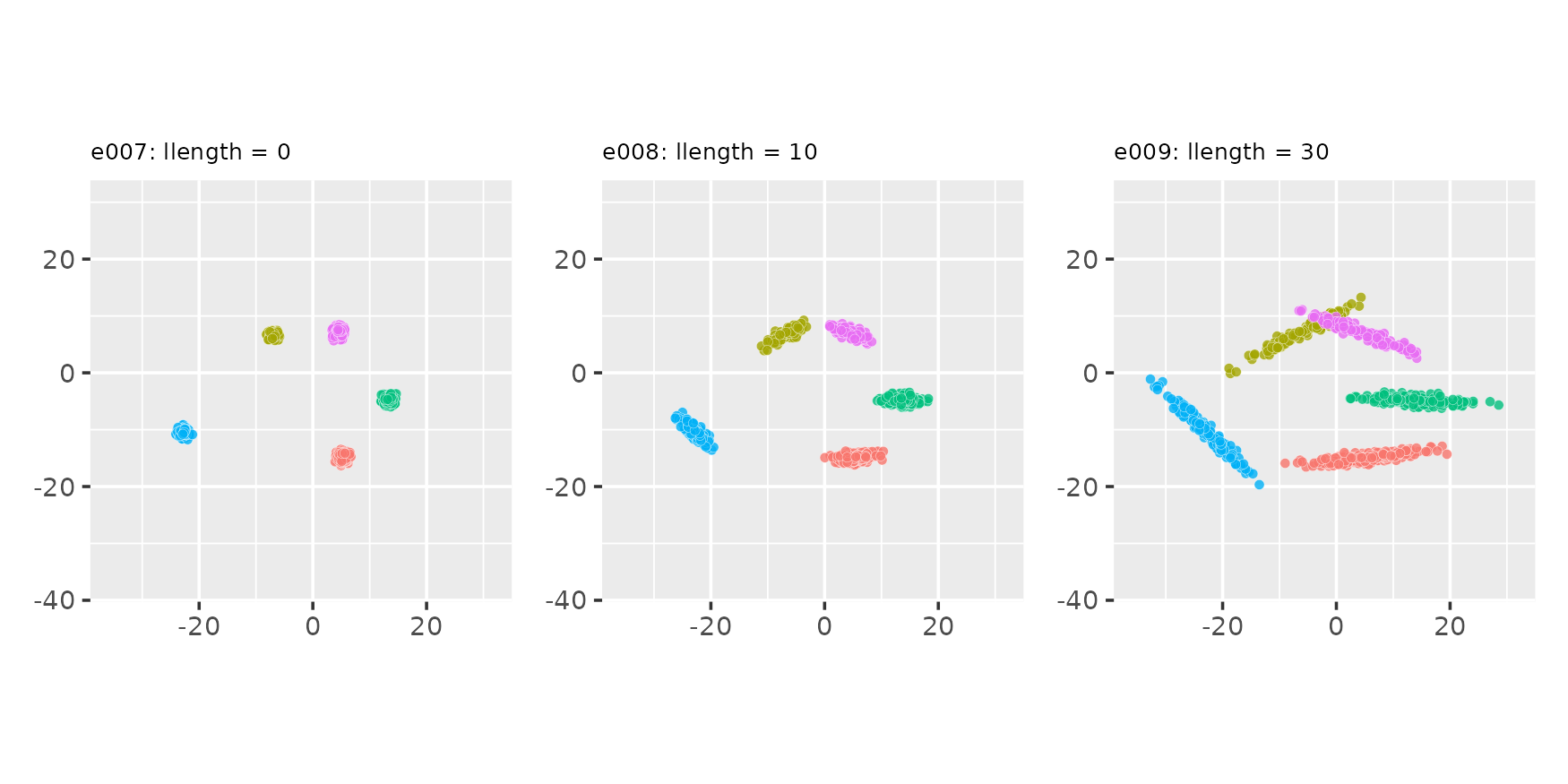
Changing the llength_disp parameter and using a custom
llengths_fn function
seed <- 765
# Custom llengths function: line lengths grow for each new cluster
llen_grow_fn <- function(nclu, llen, llenstd) {
llen * 0:(nclu - 1) + rnorm(nclu, sd = llenstd)
}
e010 <- clugen(2, 5, 800, c(1, 0), pi / 10, c(10, 10), 15, 0.0, 0.5,
seed = seed, point_dist_fn = "n")
e011 <- clugen(2, 5, 800, c(1, 0), pi / 10, c(10, 10), 15, 10.0, 0.5,
seed = seed, point_dist_fn = "n")
e012 <- clugen(2, 5, 800, c(1, 0), pi / 10, c(10, 10), 10, 0.1, 0.5,
seed = seed, llengths_fn = llen_grow_fn, point_dist_fn = "n")
plot_examples_2d(list(e = e010, t = "e010: llength_disp = 0.0"),
list(e = e011, t = "e011: llength_disp = 5.0"),
list(e = e012, t = "e012: custom llengths function"))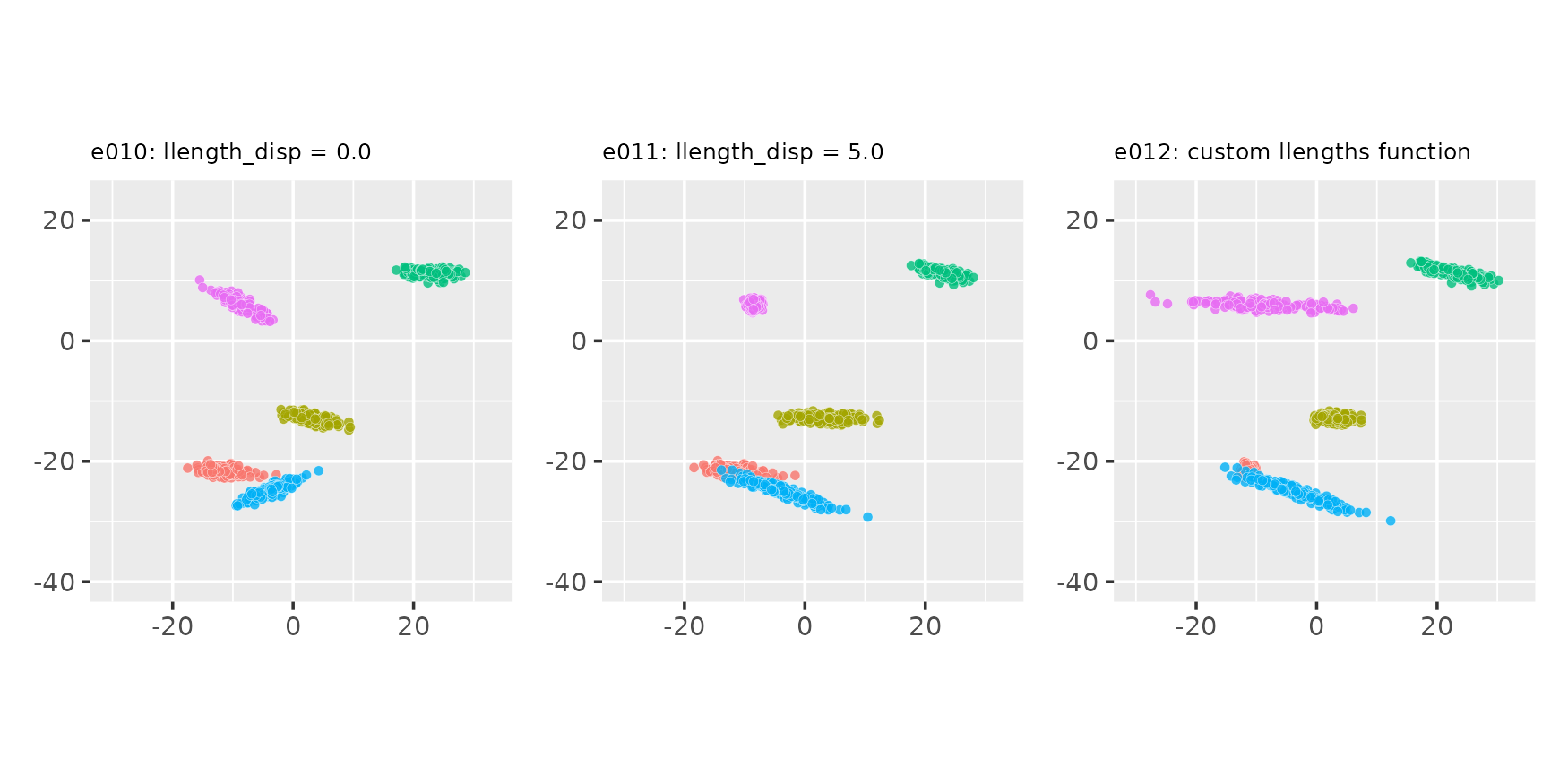
Manipulating relative cluster positions
Using the cluster_sep parameter
seed <- 222
e013 <- clugen(2, 8, 1000, c(1, 1), pi / 4, c(10, 10), 15, 2, 4, seed = seed)
e014 <- clugen(2, 8, 1000, c(1, 1), pi / 4, c(30, 10), 15, 2, 4, seed = seed)
e015 <- clugen(2, 8, 1000, c(1, 1), pi / 4, c(10, 30), 15, 2, 4, seed = seed)
plot_examples_2d(list(e = e013, t = "e013: cluster_sep = [10, 10]"),
list(e = e014, t = "e014: cluster_sep = [30, 10]"),
list(e = e015, t = "e015: cluster_sep = [10, 30]"))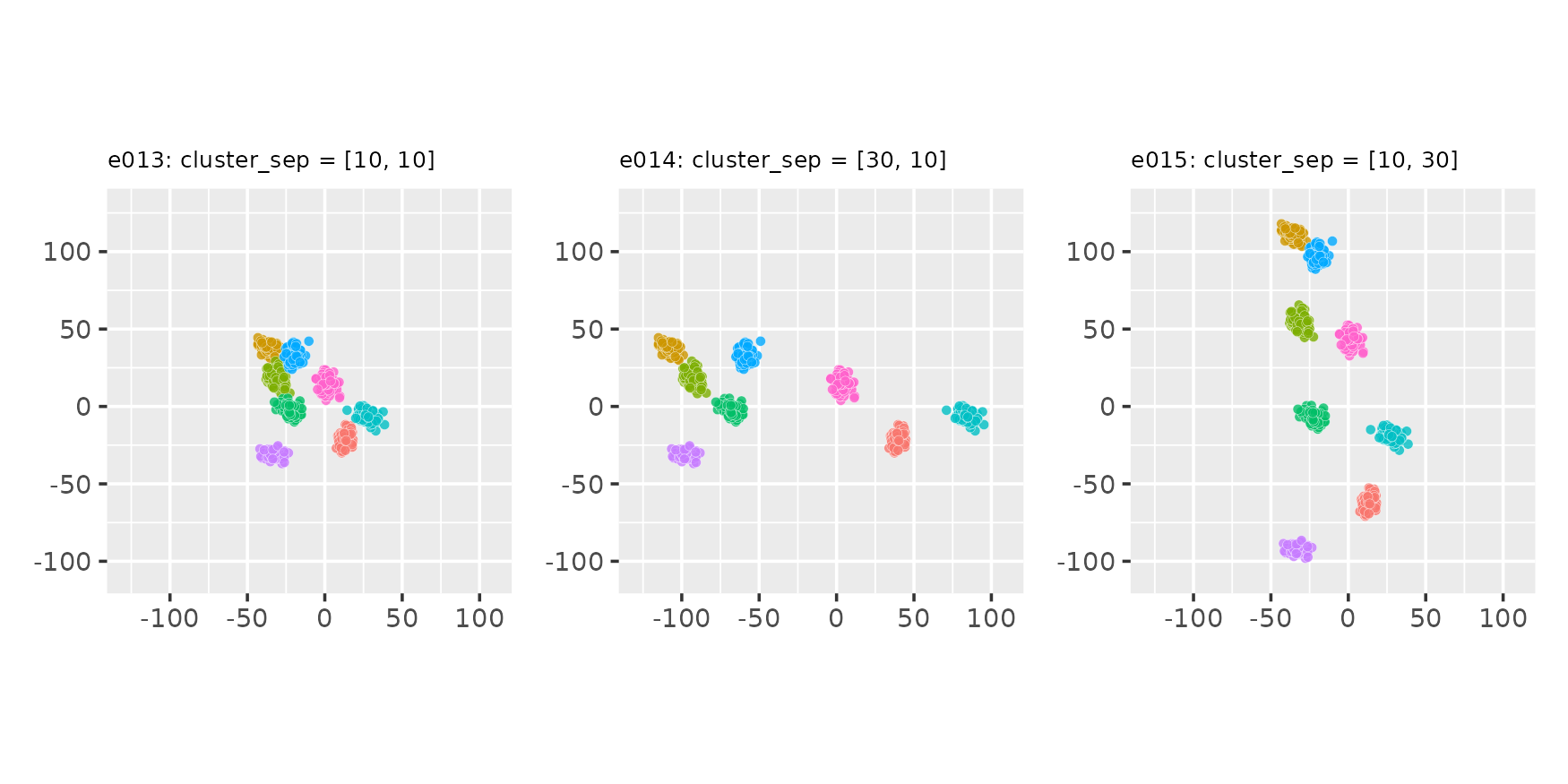
Changing the cluster_offset parameter and using a
custom clucenters_fn function
seed <- 222
# Custom clucenters function: places clusters in a diagonal
centers_diag <- function(nclu, csep, coff) {
matrix(1, nrow = nclu, ncol = length(csep)) * (1:nclu * max(csep)) +
rep(coff, each = nclu)
}
e016 <- clugen(2, 8, 1000, c(1, 1), pi / 4, c(10, 10), 10, 2, 2.5, seed = seed)
e017 <- clugen(2, 8, 1000, c(1, 1), pi / 4, c(10, 10), 10, 2, 2.5, seed = seed,
cluster_offset = c(20, -20))
e018 <- clugen(2, 8, 1000, c(1, 1), pi / 4, c(10, 10), 10, 2, 2.5, seed = seed,
cluster_offset = c(-50, -50), clucenters_fn = centers_diag)
plot_examples_2d(list(e = e016, t = "e016: default"),
list(e = e017, t = "e017: cluster_offset = [20, -20]"),
list(e = e018, t = "e018: custom clucenters function"))
Lateral dispersion and placement of point projections on the line
seed <- 234Normal projection placement (default):
proj_dist_fn = "norm"
e019 <- clugen(2, 4, 1000, c(1, 0), pi / 2, c(20, 20), 13, 2, 0.0, seed = seed)
e020 <- clugen(2, 4, 1000, c(1, 0), pi / 2, c(20, 20), 13, 2, 1.0, seed = seed)
e021 <- clugen(2, 4, 1000, c(1, 0), pi / 2, c(20, 20), 13, 2, 3.0, seed = seed)
plot_examples_2d(list(e = e019, t = "e019: lateral_disp = 0"),
list(e = e020, t = "e020: lateral_disp = 1"),
list(e = e021, t = "e021: lateral_disp = 3"))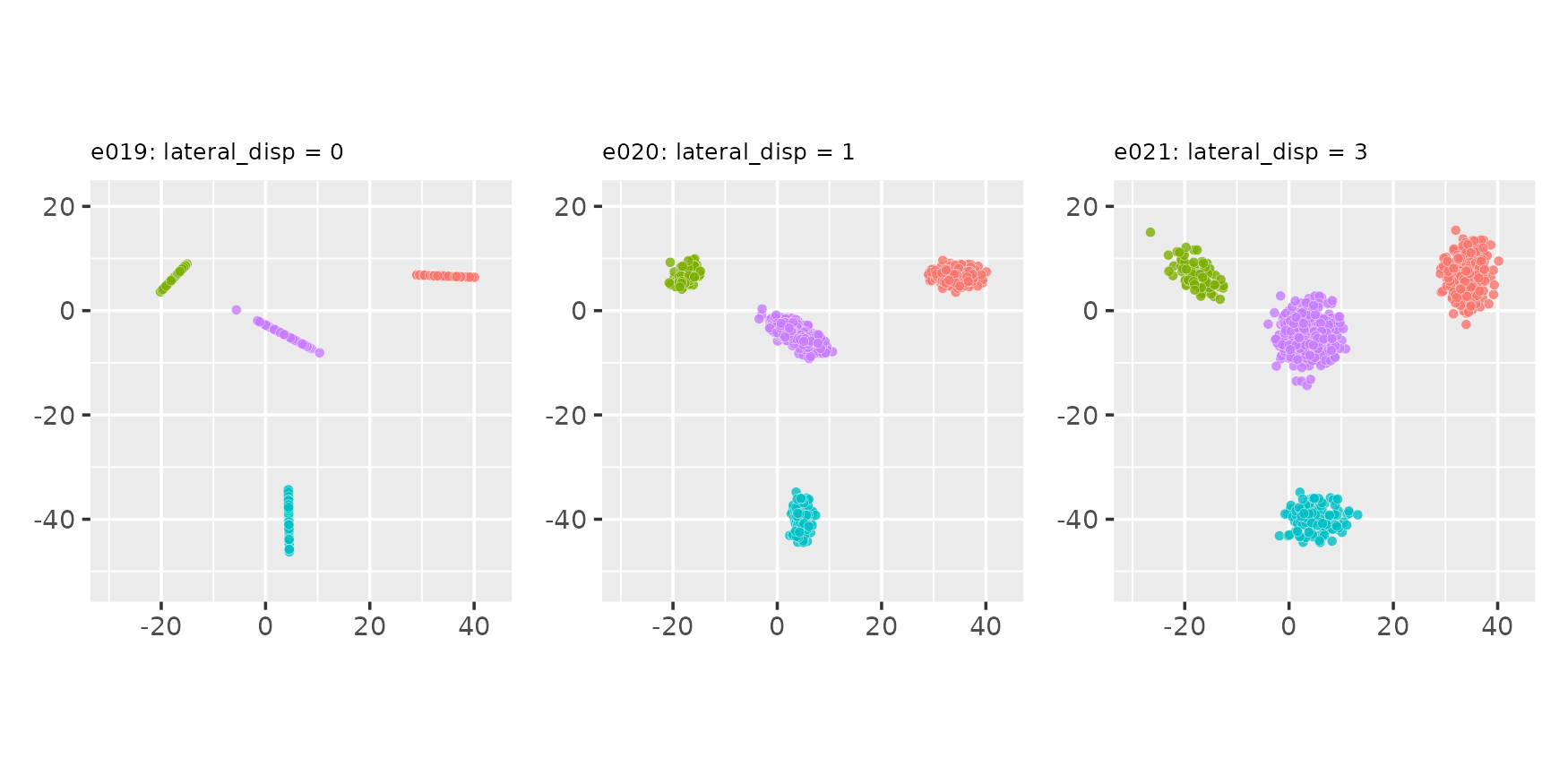
Uniform projection placement:
proj_dist_fn = "unif"
e022 <- clugen(2, 4, 1000, c(1, 0), pi / 2, c(20, 20), 13, 2, 0.0, seed = seed,
proj_dist_fn = "unif")
e023 <- clugen(2, 4, 1000, c(1, 0), pi / 2, c(20, 20), 13, 2, 1.0, seed = seed,
proj_dist_fn = "unif")
e024 <- clugen(2, 4, 1000, c(1, 0), pi / 2, c(20, 20), 13, 2, 3.0, seed = seed,
proj_dist_fn = "unif")
plot_examples_2d(list(e = e022, t = "e022: lateral_disp = 0"),
list(e = e023, t = "e023: lateral_disp = 1"),
list(e = e024, t = "e024: lateral_disp = 3"))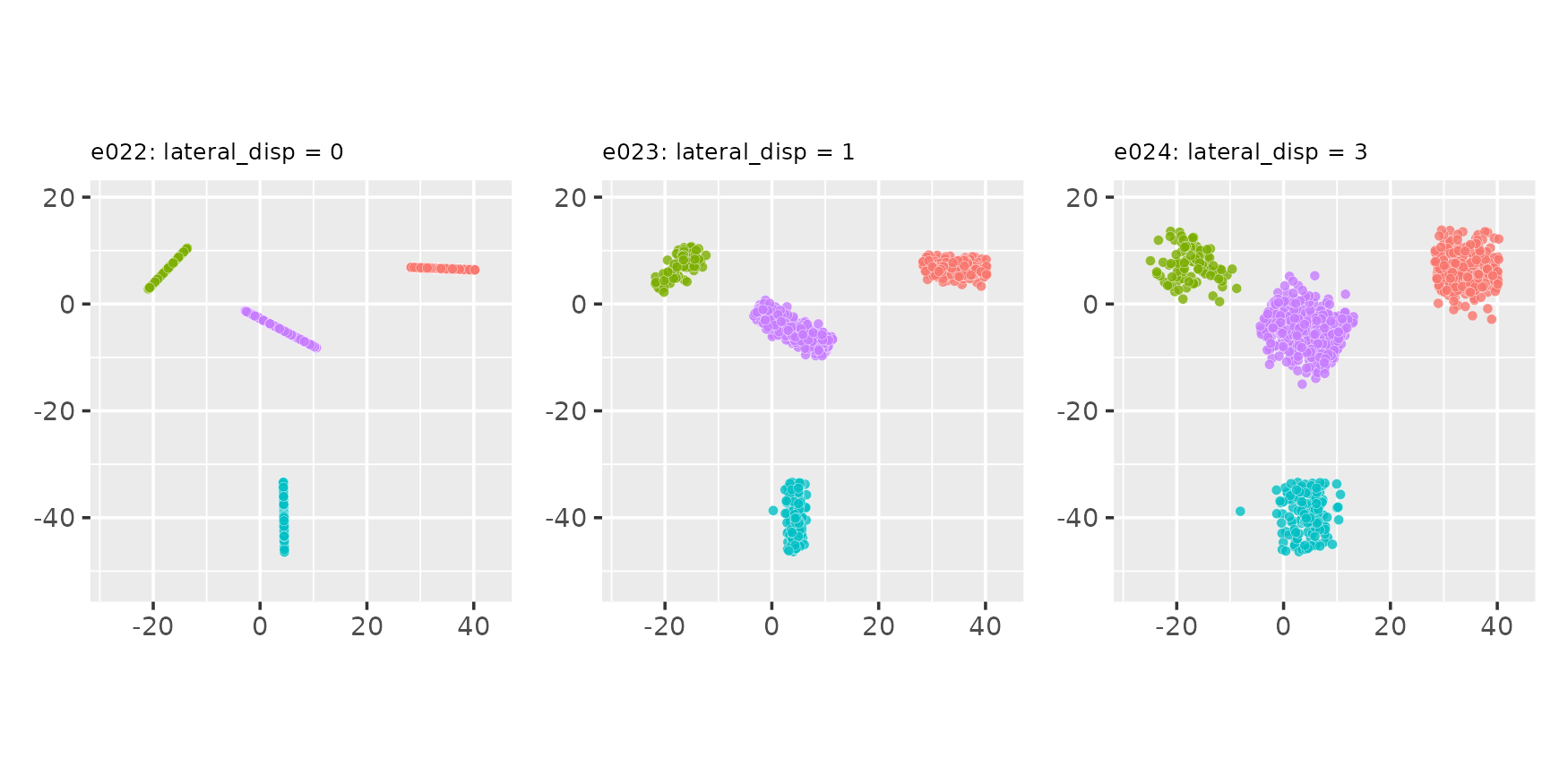
Custom projection placement using the Beta distribution
# Custom proj_dist_fn: point projections placed using the Beta distribution
proj_beta <- function(len, n) len * rbeta(n, 0.1, 0.1) - len / 2
e025 <- clugen(2, 4, 1000, c(1, 0), pi / 2, c(20, 20), 13, 2, 0.0, seed = seed,
proj_dist_fn = proj_beta)
e026 <- clugen(2, 4, 1000, c(1, 0), pi / 2, c(20, 20), 13, 2, 1.0, seed = seed,
proj_dist_fn = proj_beta)
e027 <- clugen(2, 4, 1000, c(1, 0), pi / 2, c(20, 20), 13, 2, 3.0, seed = seed,
proj_dist_fn = proj_beta)
plot_examples_2d(list(e = e025, t = "e025: lateral_disp = 0"),
list(e = e026, t = "e026: lateral_disp = 1"),
list(e = e027, t = "e027: lateral_disp = 3"))
Controlling final point positions from their projections on the cluster-supporting line
seed <- 12321
# Custom proj_dist_fn: point projections placed using the Beta distribution
proj_beta <- function(len, n) len * rbeta(n, 0.1, 0.1) - len / 2Points on hyperplane orthogonal to cluster-supporting line
(default): point_dist_fn = "n-1"
e028 <- clugen(2, 5, 1500, c(1, 0), pi / 4, c(20, 20), 14, 2, 2.0, seed = seed)
e029 <- clugen(2, 5, 1500, c(1, 0), pi / 4, c(20, 20), 14, 2, 2.0, seed = seed,
proj_dist_fn = "unif")
e030 <- clugen(2, 5, 1500, c(1, 0), pi / 4, c(20, 20), 14, 2, 2.0, seed = seed,
proj_dist_fn = proj_beta)
plot_examples_2d(list(e = e028, t = "e028: proj_dist_fn = 'norm' (default)"),
list(e = e029, t = "e029: proj_dist_fn = 'unif'"),
list(e = e030, t = "e030: custom proj_dist_fn (Beta dist.)"))
Points around projection on cluster-supporting line:
point_dist_fn = "n"
e031 <- clugen(2, 5, 1500, c(1, 0), pi / 4, c(20, 20), 14, 2, 2.0, seed = seed,
point_dist_fn = "n")
e032 <- clugen(2, 5, 1500, c(1, 0), pi / 4, c(20, 20), 14, 2, 2.0, seed = seed,
point_dist_fn = "n", proj_dist_fn = "unif")
e033 <- clugen(2, 5, 1500, c(1, 0), pi / 4, c(20, 20), 14, 2, 2.0, seed = seed,
point_dist_fn = "n", proj_dist_fn = proj_beta)
plot_examples_2d(list(e = e031, t = "e031: proj_dist_fn = 'norm' (default)"),
list(e = e032, t = "e032: proj_dist_fn = 'unif'"),
list(e = e033, t = "e033: custom proj_dist_fn (Beta dist.)"))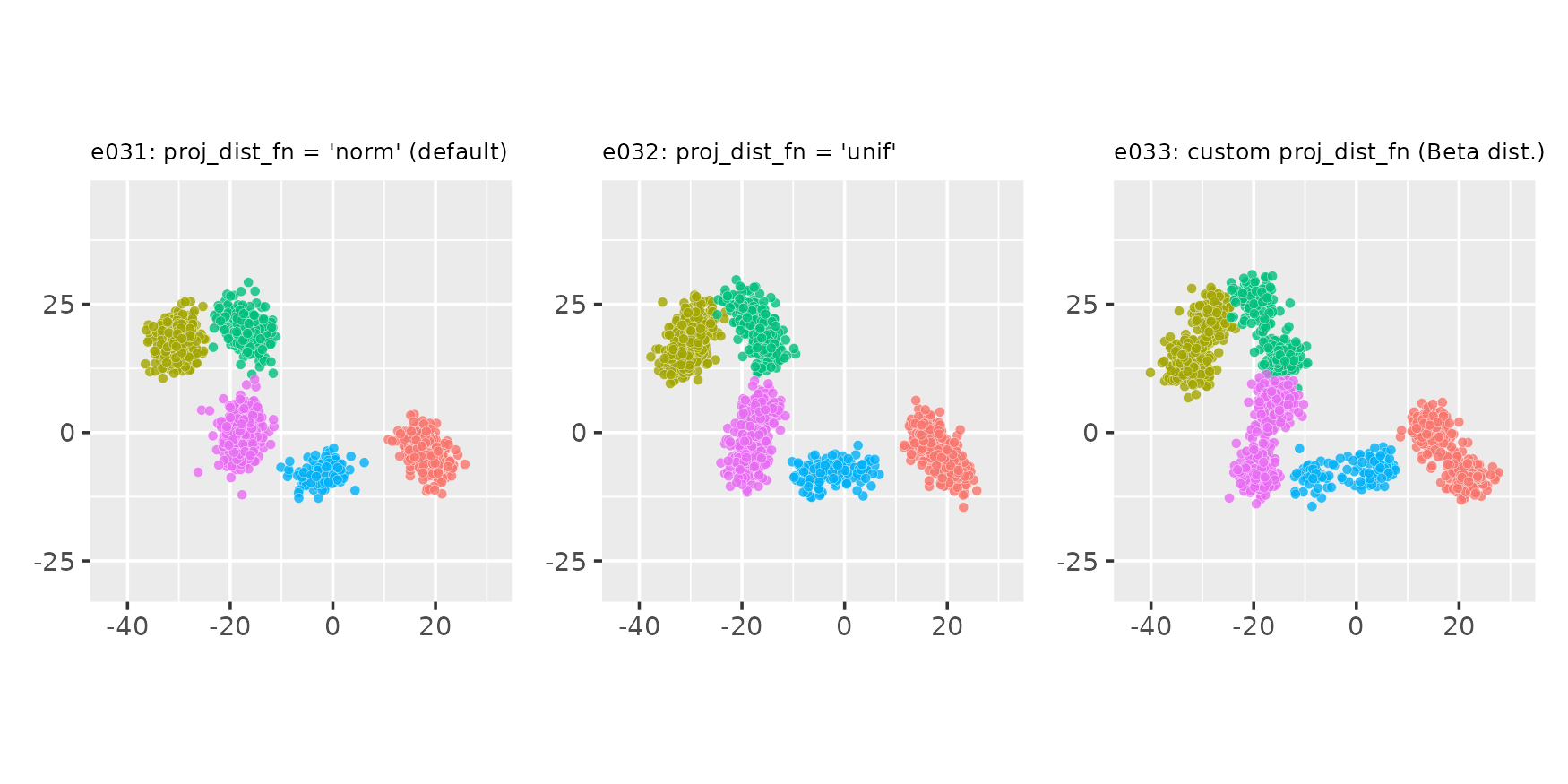
Custom point placement using the exponential distribution
# Custom point_dist_fn: final points placed using the Exponential distribution
clupoints_n_1_exp <- function(projs, lat_std, len, clu_dir, clu_ctr) {
dist_exp <- function(npts, lstd) lstd * rexp(npts, rate = 2 / lstd)
clupoints_n_1_template(projs, lat_std, clu_dir, dist_exp)
}
e034 <- clugen(2, 5, 1500, c(1, 0), pi / 4, c(20, 20), 14, 2, 2.0, seed = seed,
point_dist_fn = clupoints_n_1_exp)
e035 <- clugen(2, 5, 1500, c(1, 0), pi / 4, c(20, 20), 14, 2, 2.0, seed = seed,
point_dist_fn = clupoints_n_1_exp, proj_dist_fn = "unif")
e036 <- clugen(2, 5, 1500, c(1, 0), pi / 4, c(20, 20), 14, 2, 2.0, seed = seed,
point_dist_fn = clupoints_n_1_exp, proj_dist_fn = proj_beta)
plot_examples_2d(list(e = e034, t = "e034: proj_dist_fn = 'norm' (default)"),
list(e = e035, t = "e035: proj_dist_fn = 'unif'"),
list(e = e036, t = "e036: custom proj_dist_fn (Beta dist.)"))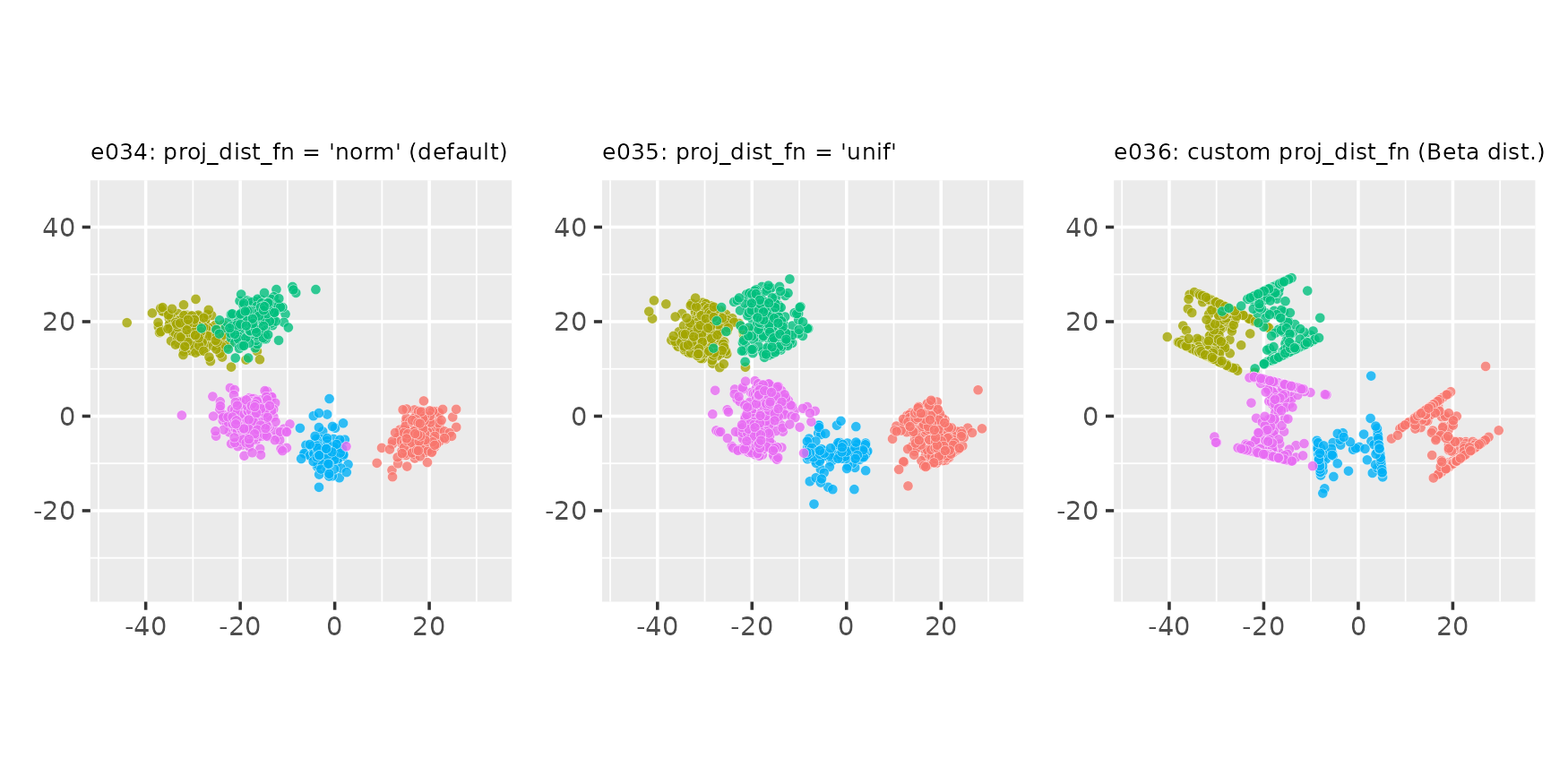
Manipulating cluster sizes
seed <- 87
# Custom clucenters_fn (all): yields fixed positions for the clusters
centers_fixed <- function(nclu, csep, coff) {
matrix(c(-csep[1], -csep[2], csep[1], -csep[2], -csep[1], csep[2], csep[1], csep[2]),
nrow = nclu, byrow = TRUE)
}
# Custom clusizes_fn (e038): cluster sizes determined via the uniform distribution,
# no correction for total points
clusizes_unif <- function(nclu, npts, ae) sample(2 * npts / nclu, nclu, replace = TRUE)
# Custom clusizes_fn (e039): clusters all have the same size, no correction for
# total points
clusizes_equal <- function(nclu, npts, ae) npts %/% nclu * rep.int(1, nclu)
e037 <- clugen(2, 4, 1500, c(1, 1), pi, c(20, 20), 0, 0, 5, seed = seed,
clucenters_fn = centers_fixed, point_dist_fn = "n")
e038 <- clugen(2, 4, 1500, c(1, 1), pi, c(20, 20), 0, 0, 5, seed = seed,
clucenters_fn = centers_fixed, clusizes_fn = clusizes_unif,
point_dist_fn = "n")
e039 <- clugen(2, 4, 1500, c(1, 1), pi, c(20, 20), 0, 0, 5, seed = seed,
clucenters_fn = centers_fixed, clusizes_fn = clusizes_equal,
point_dist_fn = "n")
plot_examples_2d(list(e = e037, t = "e037: normal dist. (default)"),
list(e = e038, t = "e038: unif. dist. (custom)"),
list(e = e039, t = "e039: equal size (custom)"))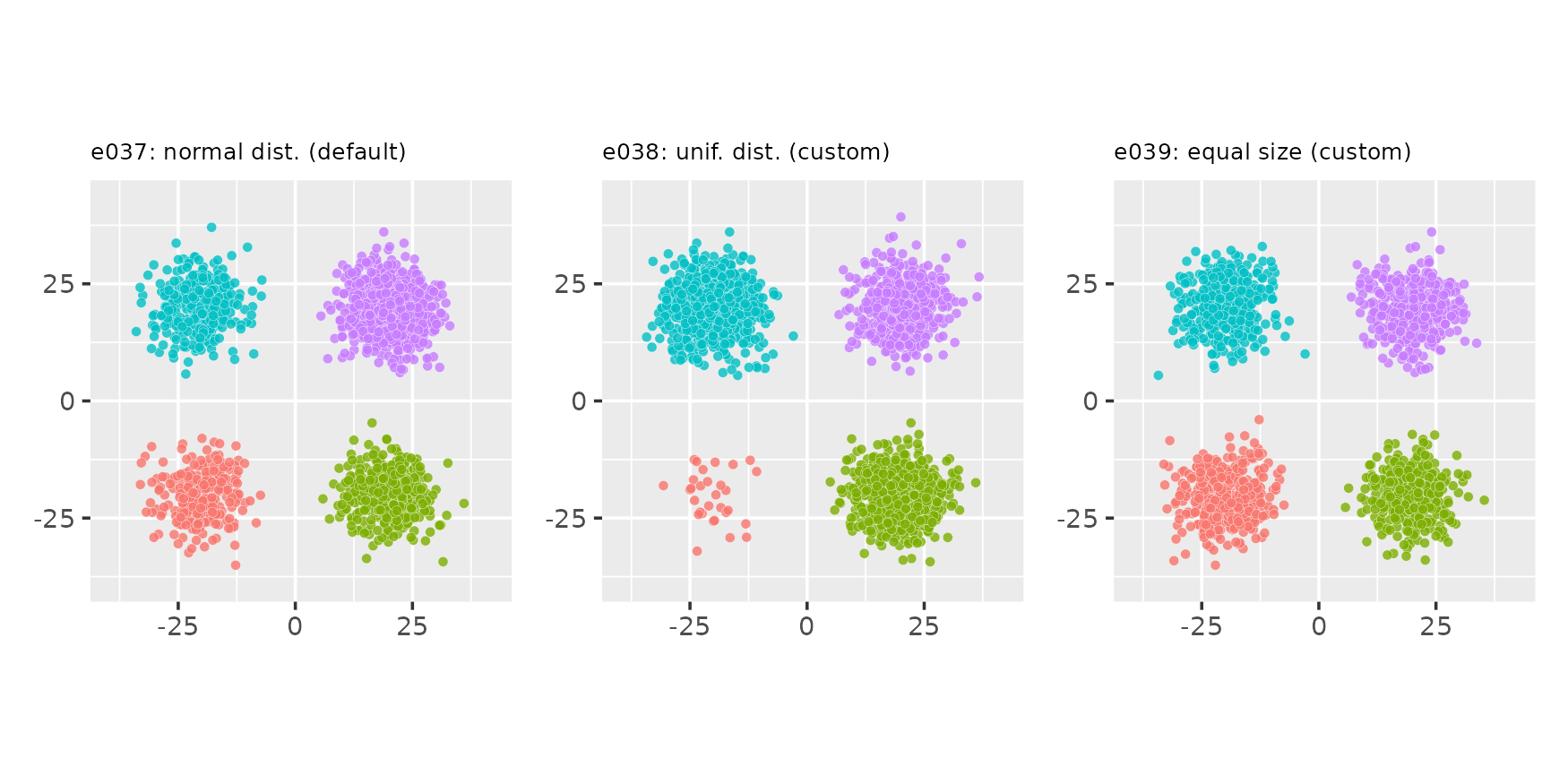
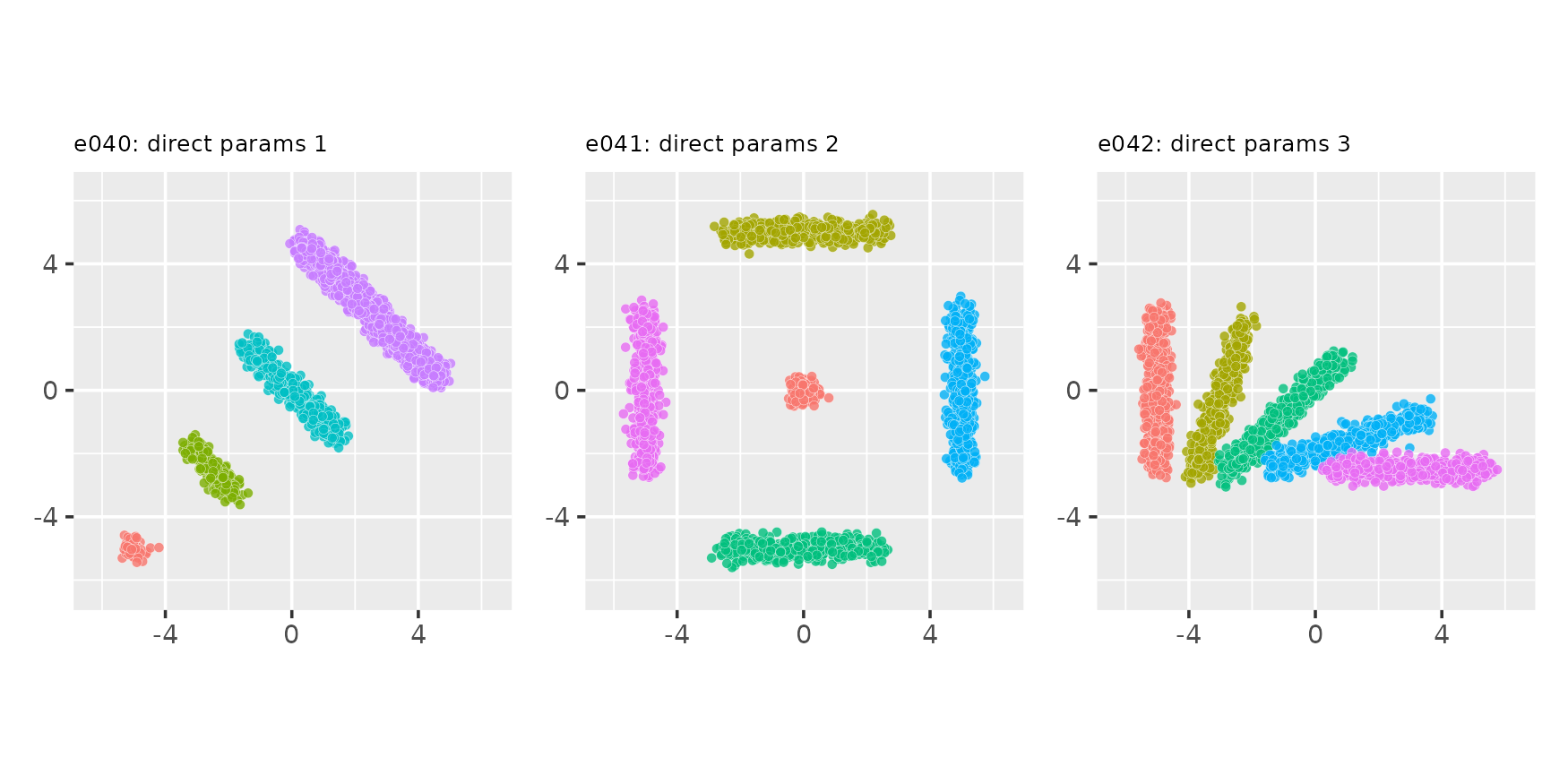
Direct specification of optional parameters
seed <- 9876
e040 <- clugen(2, 4, 1000, c(-1, 1), 0, c(0, 0), 0, 0, 0.2, seed = seed,
proj_dist_fn = "unif", point_dist_fn = "n",
clusizes_fn = c(50, 200, 500, 2000), llengths_fn = c(0, 2, 4, 6),
clucenters_fn = matrix(c(-5, -5, -2.5, -2.5, 0, 0, 2.5, 2.5),
nrow = 4, byrow = TRUE))
e041 <- clugen(2, 5, 1000, matrix(c(1, 1, 1, 0, 1, 0, 0, 1, 0, 1),
nrow = 5, byrow = TRUE),
0, c(0, 0), 0, 0, 0.2, seed = seed,
proj_dist_fn = "unif", point_dist_fn = "n",
clusizes_fn = c(200, 500, 500, 500, 500),
llengths_fn = c(0, 5, 5, 5, 5),
clucenters_fn = matrix(c(0, 0, 0, 5, 0, -5, 5, 0, -5, 0),
nrow = 5, byrow = TRUE))
e042 <- clugen(2, 5, 1000, matrix(c(0, 1, 0.25, 0.75, 0.5, 0.5, 0.75, 0.25, 1, 0),
nrow = 5, byrow = TRUE),
0, c(0, 0), 5, 0, 0.2, seed = seed,
proj_dist_fn = "unif", point_dist_fn = "n",
clusizes_fn = c(500, 500, 500, 500, 500),
clucenters_fn = matrix(c(-5, 0, -3, -0.3, -1, -0.8, 1, -1.6, 3, -2.5),
nrow = 5, byrow = TRUE))
plot_examples_2d(list(e = e040, t = "e040: direct params 1"),
list(e = e041, t = "e041: direct params 2"),
list(e = e042, t = "e042: direct params 3"))