clugen¶
Generate multidimensional clusters.
cludata = clugen( ...
num_dims, ...
num_clusters, ...
num_points, ...
direction, ...
angle_disp, ...
cluster_sep, ...
llength, ...
llength_disp, ...
lateral_disp, ...
varargin)
This is the main function of the MOCluGen package, and possibly the only function most users will need.
Arguments (mandatory)¶
num_dims: Number of dimensions.num_clusters: Number of clusters to generate.num_points: Total number of points to generate.direction: Average direction of the cluster-supporting lines. Can be a vector of lengthnum_dims(same direction for all clusters) or a matrix of sizenum_clustersxnum_dims(one direction per cluster).angle_disp: Angle dispersion of cluster-supporting lines (radians).cluster_sep: Average cluster separation in each dimension (vector ofnum_dimselements).llength: Average length of cluster-supporting lines.llength_disp: Length dispersion of cluster-supporting lines.lateral_disp: Cluster lateral dispersion, i.e., dispersion of points from their projection on the cluster-supporting line.
Note that the terms "average" and "dispersion" refer to measures of central tendency and statistical dispersion, respectively. Their exact meaning depends on the optional arguments, described next.
Arguments (optional)¶
allow_empty: Allow empty clusters?falseby default.cluster_offset: Offset to add to all cluster centers. By default the offset will be equal tozeros(num_dims, 1).proj_dist_fn: Distribution of point projections along cluster-supporting lines, with three possible values:'norm'(default): Distribute point projections along lines using a normal distribution (μ=line center, σ=llength/6).'unif': Distribute points uniformly along the line.- User-defined function, which accepts two parameters, line length (float) and
number of points (integer), and returns an array containing the distance of
each point projection to the center of the line. For example, the
"norm"option roughly corresponds to@(len, n) len * randn(n, 1) / 6.
point_dist_fn: Controls how the final points are created from their projections on the cluster-supporting lines, with three possible values:'n-1'(default): Final points are placed on a hyperplane orthogonal to the cluster-supporting line, centered at each point's projection, using the normal distribution (μ=0, σ=lateral_disp). This is done by theclupoints_n_1()function.'n': Final points are placed around their projection on the cluster-supporting line using the normal distribution (μ=0, σ=lateral_disp). This is done by theclupoints_n()function.- User-defined function: The user can specify a custom point placement strategy
by passing a function with the same signature as
clupoints_n_1()and `clupoints_n().
clusizes_fn: Distribution of cluster sizes. By default, cluster sizes are determined by theclusizes()function, which uses the normaldistribution (μ=num_points/num_clusters, σ=μ/3), and assures that the finalcluster sizes add up tonum_points. This parameter allows the user to specify a custom function for this purpose, which must followclusizes()signature. Note that custom functions are not required to strictly obey thenum_pointsparameter. Alternatively, the user can specify a vector of cluster sizes directly.clucenters_fn: Distribution of cluster centers. By default, cluster centers are determined by theclucenters()function, which uses the uniform distribution, and takes into account thenum_clustersandcluster_sepparameters for generating well-distributed cluster centers. This parameter allows the user to specify a custom function for this purpose, which must followclucenters()signature. Alternatively, the user can specify a matrix of sizenum_clustersxnum_dimswith the exact cluster centers.llengths_fn: Distribution of line lengths. By default, the lengths of cluster-supporting lines are determined by thellengths()function, which uses the folded normal distribution (μ=llength, σ=llength_disp). This parameter allows the user to specify a custom function for this purpose, which must followllengths()signature. Alternatively, the user can specify a vector of line lengths directly.angle_deltas_fn: Distribution of line angle differences with respect todirection. By default, the angles betweendirectionand the direction of cluster-supporting lines are determined by theangle_deltas()function, which uses the wrapped normal distribution (μ=0, σ=angle_disp) with support in the interval [-π/2, π/2]. This parameter allows the user to specify a custom function for this purpose, which must followangle_deltas()signature. Alternatively, the user can specify a vector of angle deltas directly.seed: Non-negative integer for initializing the PRNG, allowing for reproducible results; alternatively, the PRNG can be initialized with thecluseed()function, or by directly setting the seed in MATLAB or Octave (each using its own specific approach).
Return values¶
A struct with the following fields:
points: Anum_pointsxnum_dimsmatrix with the generated points for all clusters.clusters: Anum_pointsx 1 vector indicating which cluster each point inpointsbelongs to.projections: Anum_pointsxnum_dimsmatrix with the point projections on the cluster-supporting lines.sizes: Anum_clustersx 1 vector with the number of points in each cluster.centers: Anum_clustersxnum_dimsmatrix with the coordinates of the cluster centers.directions: Anum_clustersxnum_dimsmatrix with the direction of each cluster-supporting line.angles: Anum_clustersx 1 vector with the angles between the cluster-supporting lines and the main direction.lengths: Anum_clustersx 1 vector with the lengths of the cluster-supporting lines.
Note that if a custom function was given in the clusizes_fn parameter, it is
possible that num_points may have a different value than what was specified in
clugen's num_points parameter.
Note¶
This function is stochastic. For reproducibility use the seed parameter or
set the PRNG seed as discussed in the Reference.
Examples¶
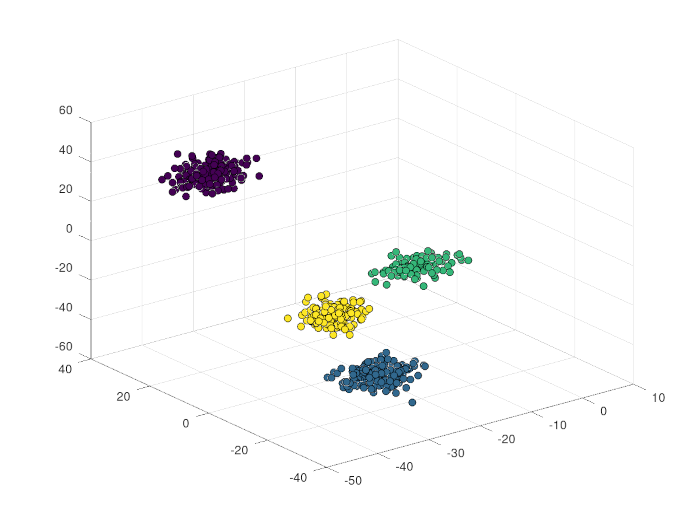
This creates 4 clusters in 3D space with a total of 1000 points, with a main
direction of [1; 0; 0] (i.e., along the x-axis), with an angle dispersion of
pi / 8, average cluster separation of [20; 15; 25], average length of
cluster-supporting lines of 16 (dispersion of 4 units), and lateral_disp of
3.5. The seed parameter is set to 123, demonstrating how to use the optional
arguments.
The following command plots the generated clusters:
scatter3(o.points(:, 1), o.points(:, 2), o.points(:,3), 36, o.clusters, 'filled', 'MarkerEdgeColor', 'k');